
Golang and DNA Synthesis
The intersection of human biology and technology is ever-evolving. As software solutions become more and more integrated into everyday life (looking at you, Alexa) and AI and machine learning continue their rapid trajectory towards 21st-century dominance, it’s becoming harder and harder for one to exist without the other. And that’s not necessarily a bad thing.
In fact, it’s technology that is powering the current testing for SARS CoV-2, the novel coronavirus responsible for the pandemic causing a widespread pneumonia-like illness called COVID-19. PCR, or polymerase chain reaction, is a standard bio-molecular testing technique that uses small snippets of DNA to detect extremely tiny amounts of virus in a specimen, i.e., a nose or throat swab. Read on to learn more about the technique as well as its application in testing for SARS CoV-2.
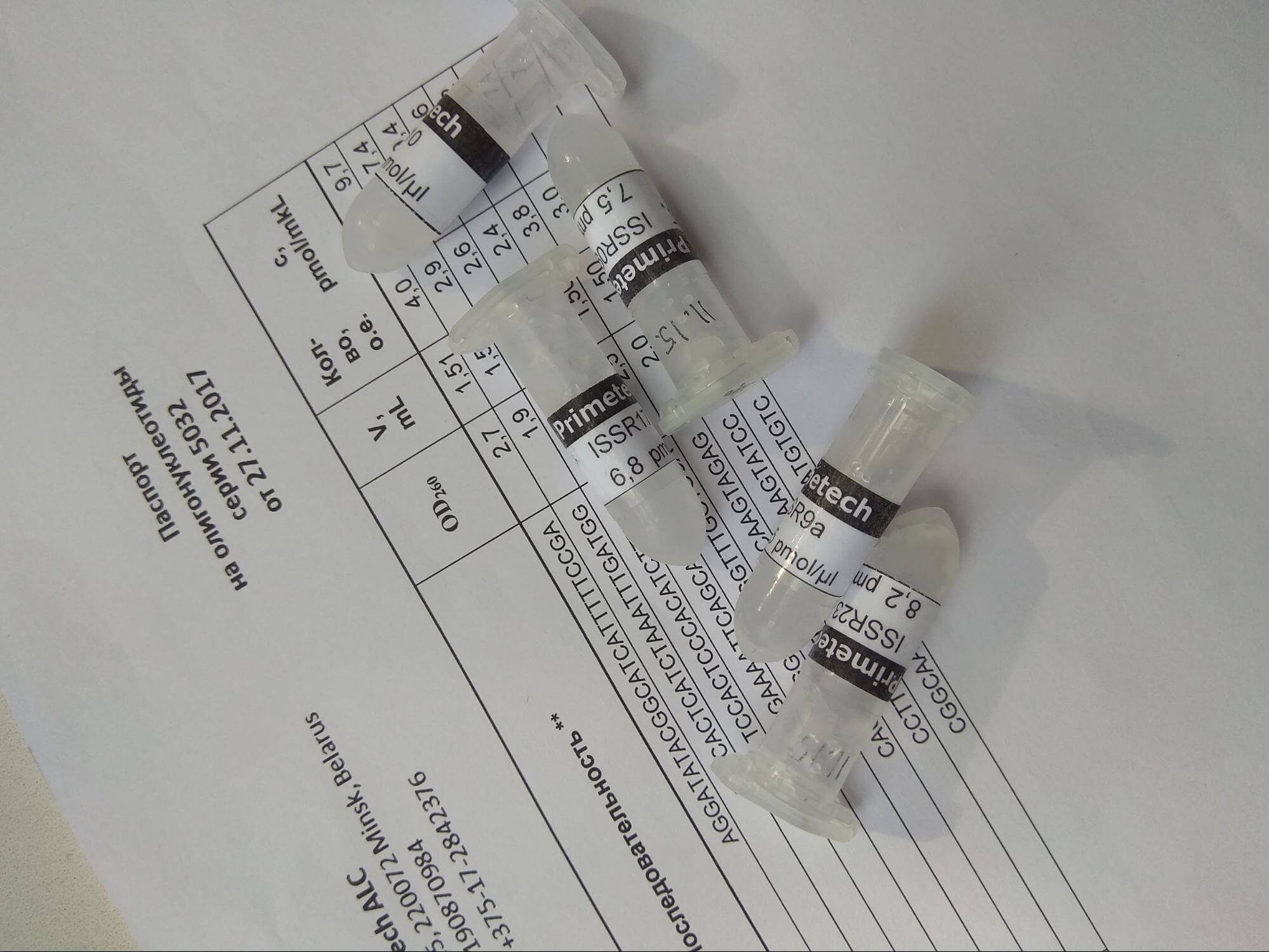
I would like to share my experience from working for a small company in the biochemistry domain. The company produces short DNA fragments called oligonucleotides or primers. To give you an idea of the size of primers, on average, a strand of DNA will consist of hundreds of thousands of “links.” The fragments of DNA that we produce typically contain only 40 “links.” In a month, we output roughly 800 primers which are used for various types of analyses.
That output isn’t necessarily high compared to other primer producers, however, I am always looking for interesting ways to optimize the process and make it more fun. I decided to explore some of my ideas and record statistics on the production of DNA by using the Golang (Go) programming language. You're likely already wondering, “Who needs these primers?”, “What are DNA sequences?”, or “Why Golang?”. I'll start from the beginning.
DNA and RNA - Hard Drive and RAM
In modern computing, a binary coding system is used for storing information. Multiple alternations of "0" and "1" allow you to save and transmit text, photos, videos, etc. In the cells of living organisms, DNA (Deoxyribonucleic acid) becomes the coding system, but instead of a binary implementation, information is stored using a quaternary system–that is, DNA strands consist of millions of combinations of four chemical units.
Chains of DNA molecules are interconnected in pairs, forming a strong structure in which to store information for long periods of time (under normal conditions). This information is like a kind of biochemical hard disk of living cells that provides the genetic instructions for how organisms will develop, grow, function, and reproduce. And of course, there is a mechanism for replicating the necessary information in each cell.

The RNA (Ribonucleic acid) chain also consists of the alternation of four chemical units, though they differ slightly from the chemical makeup of the DNA units. In living cells, RNA forms an image of the DNA strand, while the DNA strand itself remains unchanged (information continues to be stored on the hard disk). But the RNA chain (a chemical cast from a cell's hard disk) is already a team for a living cell to produce the necessary proteins. Therefore, a living organism protects its cells from the ingestion of foreign RNA. That is why the production of RNA molecules is more complicated, needed less often, and it is more expensive and dangerous.
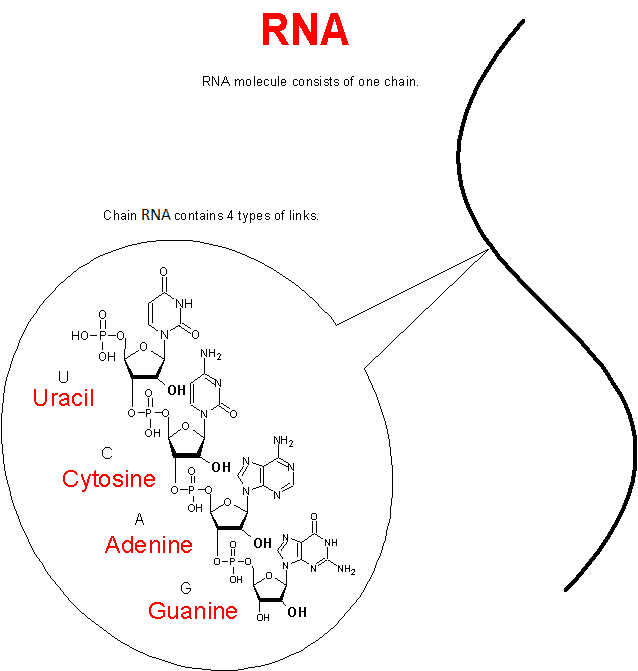
So the chemical units of the DNA molecule are only 4:
- A (adenine)
- C (cytosine)
- G (guanine)
- T (thymine)
As we see, they have a letter designation and a common name: Amidites. The DNA sequence itself has the form:
5’-AGCCGCCTAAG-3’
This is an example of a sequence of 10 links. 3’ and 5’ are symbols for the beginning and end of a sequence. Reading and writing a sequence is done from right to left from 3’ to 5’.
Other Latin letters may be present in the sequence
R, Y, K, M, S, W, B, D, H, V, N
These letters mean that at the stage of joining a new link in the chain, the device feeds a mixture of amidites (the letters of the notation are standard and are readable by any automatic DNA synthesizer). For example, R is 50% A + 50% G. With an arbitrary sequence of 5’-AGCR-3 ’as the final product, we obtain a mixture of primers 50% 5’-AGCA-3’ + 50% 5’-AGCG-3 ’.
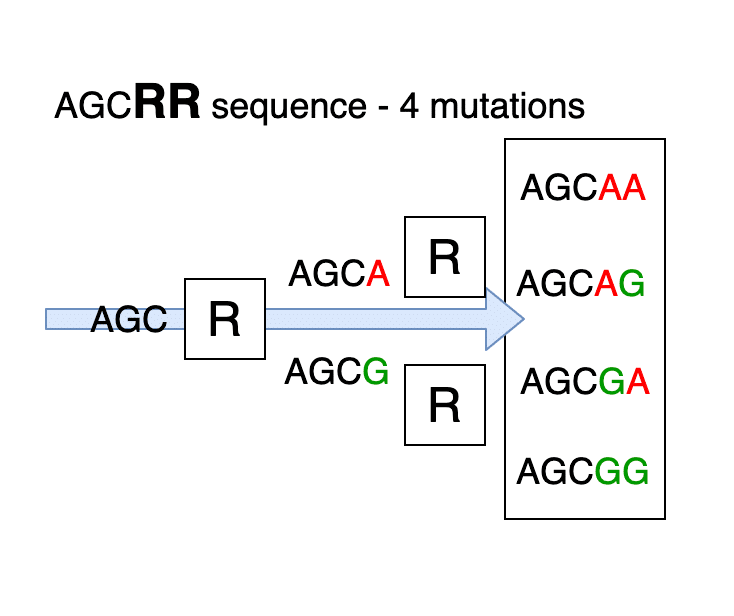
Such primers are used to detect possible mutations of DNA. The production of primers can use other modifying additives. These additives are used there is no need to know during the synthesis, but it is necessary to take into account this possibility in the program.
What are Primers For and Who Orders Them
Primers are mainly used to detect the presence of a specific type of DNA. It might be virus DNA, bacteria, a genetically-modified product, cancer cells, etc. Remember, DNA is unique to the organism from which it came. The small DNA sequences that make up the primers we deal with are determined by scientists separately.
The fact of the matter is, it is not possible to detect specific DNA in a small sample because its concentration is too low. But, if you make millions or even billions of copies of these very small amounts of DNA sequences, you are able to enlarge the concentration enough so that it can be studied in detail. The very process of DNA copying is called PCR (polymerase chain reaction) after the name of the substance polymerase that allows this process to take place. PCR
The primers produced at our laboratory isolate the segment of DNA to be copied and allow for enlarging concentration of only this fragment. The customers for whom we produce primers range from medical centers to research institutes to enterprises of the food industry.
Here is a real-life example: A medical center needs to determine whether a patient is ill with, say, a certain viral blood disease. A blood sample is taken from the patient. The task is to find out if the virus causing the disease is present in his blood or not.
The medical center charged with treating the patient reaches out to our company and asks us to make primers capable of uncovering the presence of this fragment of virus DNA. The specialist of the medical center carries out the PCR process, analyzes the PCR product, and sees that the copying did not work. That's right, it means that there was no virus in the original sample, i.e., there was nothing to copy. The doctor looks at the result of the analysis and reports the good news to the patient.
The PCR method is one of the primary high-precision methods of analysis for the novel coronavirus, SARS CoV-2. You can read more about how SARS CoV-2 testing works here. DNA fragments for analysis on COVID-19 are of the utmost urgency and our team is tasked with completing these tests immediately and in the shortest possible time.

Description of the Production Process
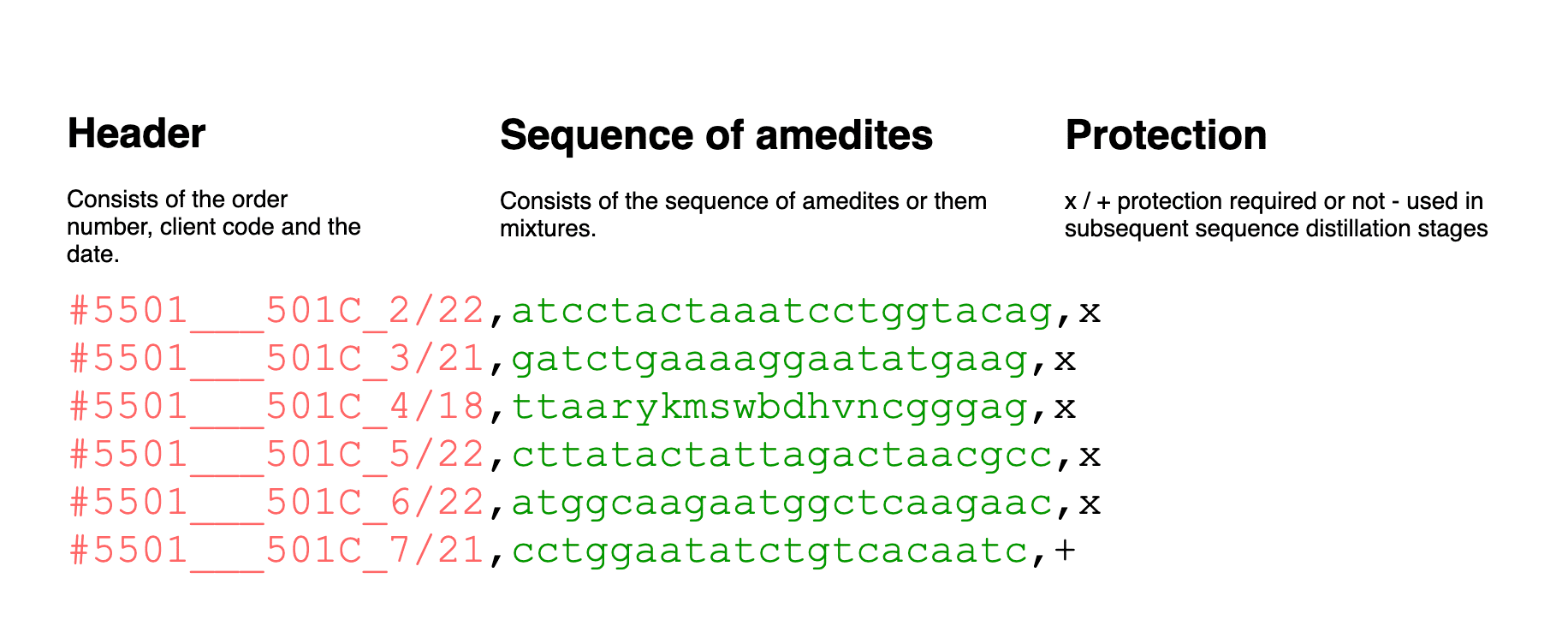
An order comes to the company either via email or through the order form on the website. The order contains a list of primers with individual names and sequences of amidites, in addition to information about the customer. The manager receives the order, copies it, and with the help of MS Excel macros, translates it into a convenient form for our work. Then it assigns an order number and saves the Excel file.
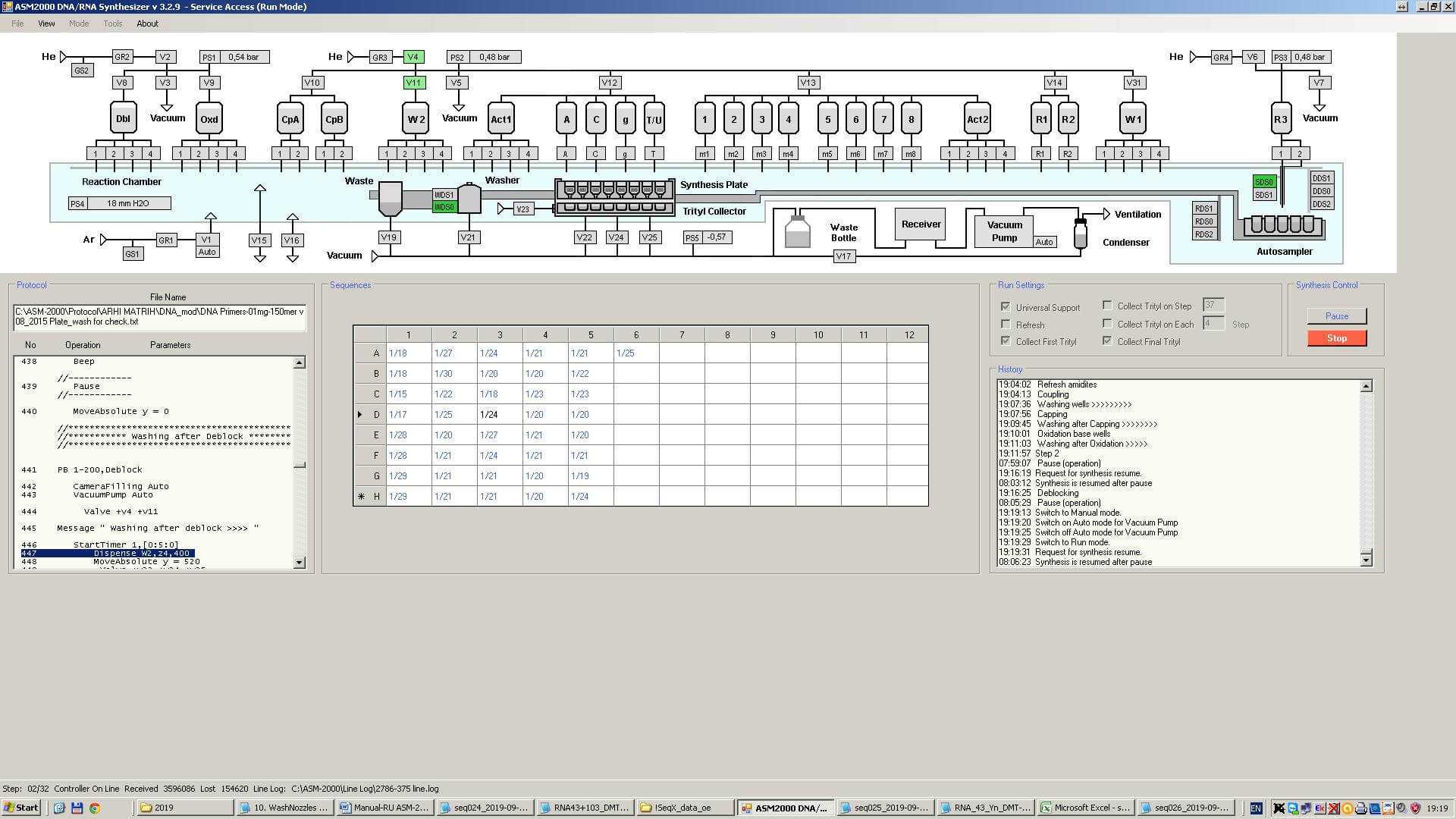
DNA synthesis is carried out using an automatic synthesizer. The synthesizer is connected to a computer with a number of reagent tanks containing amidites and other substances. A balloon with inert gas helium is connected to the synthesizer itself (the reagents are “afraid” of oxygen). All reagents are pre-dried from water because the reaction does not proceed in the presence of water.

The synthesis is carried out in a microbiological plate with 96 cells for the synthesis of 96 different sequences at the same time. It can make up to 96 different sequences simultaneously. The length of each DNA sequence varies from 5 to 80 amidites with a median of 40.
The person performing the synthesis makes up the RUN-block for the synthesizer using the “for synthesis” tab from the MS Excel files generated by the manager. RUN-block, most often, includes sequences from different orders. RUN-block is a file in TXT format:
After installing the tablet, loading the RUN-block, and checking the operability of the device, the operator selects the scale of synthesis. It is quite an important setting since it captures the amount of DNA to be obtained. Finally, she presses the start. The duration of the synthesis depends on the length of the primers and the characteristics of the printer. The printer usually takes 5-6 hours.


After the synthesis is complete, the primers need to be cleared of synthesis byproducts. These are mostly incomplete chains (the efficiency of joining each of the links of the chain is less than 100%).
Then, each of the primers is placed in a separate tube, and their quantities are measured.
They are dried, signed, and given to the customer with their order passport. The order passport indicates the date of synthesis, the names of the primers, their sequence, and the number of units in each primer.
And that’s it! The primers are ready to use.
Golang Experiment with DNA-Beaver
Why Golang
At school, I studied the programming language Pascal, and I quite liked it. Many years have passed since then, however, and the world of programming languages has changed significantly. Pascal is not considered relevant today, although, of course, it is still used.
I wanted to update my programming knowledge and try to write a program in a modern language (while also making the program simple, convenient, understandable, and full-featured). I discussed the idea with my brother, who is a programmer, and he recommended that I look at Golang.
After reading about Go, I was not disappointed. What I liked:
- The functionality and minimalism of the language as well as strict data typing; that is, the compiler and integrated development environment will always alert you to where there is an error in the code causing a data incompatibility;
- The possibility of an easy start for a beginner–the code is easy to read and understandable, the syntax is very transparent and unambiguous;
- It allows you to write full-fledged applications used in well-known projects, such as Docker for example;
- Go has everything you need to work right out of the box - they can compile the project, build dependencies, test;
- It has a garbage collector which means I do not need to worry about deleting data;
- Go has very good parallel data processing which provides ample opportunity for optimizing a program, although in my program I used this opportunity not entirely for the intended purpose, but more on that later;
- Go programs are compiled for different operating systems; they can be run on MacOS, Windows, Linux, and even Raspberry PI and Arduino.
The list goes on, but that is enough for now.
What am I going to solve with my code?
In part, I wanted to refresh my knowledge of programming, and I was interested in the following technologies:
- Golang programming language
- SQL database
- Concurrent calculations
- version control system (GIT) for saving updates on the code
What purpose does the program serve?
I would like to save the data about the orders and search for coincidences of oligos. They rarely happen, but it would be nice to have the statistics and see if they are from the same company and what they shared in common.
Also, it is always nice to have validations for the sequences before starting the process of synthesis with the DNA printer. So, my Golang program will do that.
It will be really helpful if it can calculate the amount of every amidite and send the exact right amounts to the printer (without spare and losing money). This was also included in the scope of the first version.
How to write the code
Nowadays, nobody writes code in notebooks or text editors. Instead, developers use Integrated Development Environments (IDEs). The environment helps to navigate a software project, run it, save it to the versioning systems, and many other tasks. There are a host of different IDEs: Eclipse, JetBrains, Vim, etc. I have chosen the product of JetBrains - Golang, and have found it very convenient and helpful.

Versioning Control Systems
Since I am not a first-time internet user, I decided to keep my code in the cloud so it will be accessible from anywhere and safe in the event of a local data loss. But is does there exist a cloud just for code?
Indeed! And what’s more - this type of cloud provides for simultaneous contributions and changes to the same pieces code by many developers. One of the most popular providers of this type of service is GIT https://github.com. Developers can put their code on Github and share their findings with the wider world! That is really nice! GIT can be installed easily and then used from the JetBrains Golang IDE.
I created an account on Github and made a repository for my Golang DNA experiments: https://github.com/novikov1981/dna-beaver
Then I made a folder to store all my code locally:
git checkout https://github.com/novikov1981/dna-beaver.git
You can use this set of commands for Git https://git-scm.com/doc or popup menus from Golang to save, upload, and see differences in your code versions. After the first commit, the code is made visible to the whole world since the project is public. Anyone can come in and clone your code to reuse it for their own purposes, but every time you update the code, it saves the difference–this is called version control. Version control systems like Github are essential tools for working in teams.
Installing Golang
It is quite simple to install Golang on almost any platform: https://Golang.org/doc/install#install
You can even run code snippets online without downloading Golang at all:
Structure of the project
The code consists of several folders called “packages” in the Go language:

The cmd folder contains the runnable main package with the application.
The project also contains test files in the data samples folder.
The repository package consists of the working code in the database. This package will be described later in the section titled “Work with the database."
The measurements and validations folders are for gauging functionality; they measure the amount of every amidite in the synthesis and verify if the structure of the input file and sequences consists of the correct symbols.
Interface with the user
There are a number of interfaces that work with the user, including:
- the console
- native system interfaces like MS Windows desktop or Gnome in Linux
- web interfaces working with the user within the browser

In this example, I used the standard Golang library flag. I’ll share how the flags are set to run the application in different modes later on.
Install dna-beaver
First of all, you need to have Git already installed in your computer because Golang uses it under the hood when working with dependencies. Golang modules will resolve the correct version of the libraries you are using to build your project.
Dna-beaver requires you to set an environment variable:
export GO111MODULE=on


Then you can install it in two ways:
Get it:
go get -v github.com/novikov1981/dna-beaver/cmd/dnabeaver

Or build it:
go build -o dnabeaver dna-beaver/cmd/dnabeaver/main.go

If you want to work with the source code, you can check out the whole project.
git checkout https://github.com/novikov1981/dna-beaver.git
Right after the installation of the utility, you can download the data samples from https://github.com/novikov1981/dna-beaver/tree/master/datasamples to the same location.
Run dna-beaver
The dna-beaver application’s CLI (command-line interface) consists of a number of commands with different sets of flags. All that is just out-of-the-box installation for Golang, package “flag”:

So we can check the flags for all of the commands:

The application uses the synthesis.db file in the same folder where we run the executable. If the file does not exist, it creates a new one on the first run.

Working With the Database
One of the aspects I was going to cover with this tool is the persistence of the data for the synthesis. While sometimes it is possible to save the data into files with any arbitrary format, in this case, it is necessary to write the code for saving and reading the data (which makes it more difficult to search and maintain).
Hopefully, engineers do not feel the need to reinvestigate such widely-used tools. There are a lot of databases that cover exactly this functionality - storing and searching the data. We have quite a structured set of data with fixed fields; thus, we will use a relational or SQL database.
Relational databases use SQL language to create tables, insert or update the data in the tables, and then search the tables for the data. This is exactly what we need!
The SQL databases may be really big and complicated, like PostgreSQL or OracleXE. Such complex solutions require a separate server to store the data. The DNA-Beaver project does not require this type of sophisticated solution, so I have chosen SQLite database. It stores the data in a single file, requires no server, and can be accessed just using the library from your application. I used github.com/jmoiron/sqlx library for Golang in order to work with SQLite.
Before working with the database, we planned the schema of the data:

The schema is quite simple. We can keep many synthesizes in our database, and every synthesis can contain many oligos. The data types for the fields in SQLite are quite primitive, so we used TEXT for storing created_at, which is actually the date. For our application, this level of abstraction was enough.
The database layer is located in the repository/sqllite folder and is represented as a Repository structure. This allows it to be replaced with some other database implementation later.
The repository has SQL statements for making a new database, adding new synthesis and, searching for them by name or oligo pattern.
SQLLite allows us to search by pattern %LIKE%, so we can find any oligo that includes a pattern with symbols at the beginning and the end.
Concurrency
I read a lot about concurrency in Go, and it proved to be one of the biggest advantages of the language. Concurrency allows a program to make computations in parallel, using different cores of the processor, which improves performance.
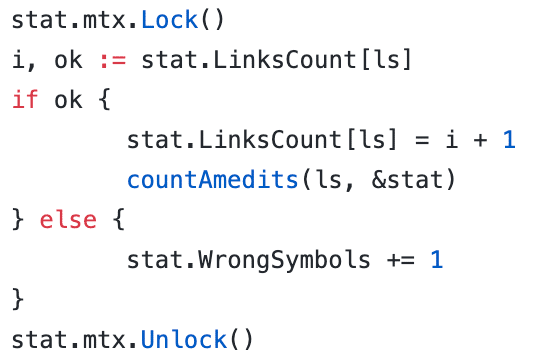
While the performance of the DNABeaver application wasn’t really an issue (because the job simply wasn’t that heavy), I still decided to tweak it and add some parallelism in the measurements of how many links we have and how many oligos are consumed with the synthesis: https://github.com/novikov1981/dna-beaver/blob/master/measurements/statistics.go
In that example, I was trying to parallelize calculations on different sequences. But, in my algorithm, I use the map. Golang map does not allow for concurrent writing to the map; therefore I had to use mutexes for the map writings.
The final performance of such parallelism is not much better than the subsequent version. So, I plan to change the algorithm not to use the map for data synchronization by channels or revert back to the consequent version.
Conclusions
In this article, I have described my first programming experience with Go. I have created a simple (for professionals, not for me) application to validate, save, and search synthesis data–an application I now use in my daily work with DNA synthesis. While I have yet to find any coincidence of DNA produced: every time we make a new sequence. But I still hope to have the same sequence one day. I have found some validation errors, however, and, ultimately, have made my working time more efficient.
It was really astonishing to see the modern tools available for creating this type of integrated development environment (IDE). Moreover, it is strange to hear that simple text editors like Vim are becoming “fashionable” nowadays. Real IDEs, like Golang, are extremely convenient, useful, and fast, but they are rarely free of charge.
Also, it is nice to have my code stored (forever, I hope!) at github.com where it can easily be shared and where people can reuse my findings if they are so inclined.
I tried to work with SQL with quite a success; at least my sequences can be saved and then found.
My experiment with concurrency showed to me that not every algorithm can be parallelized. Better to say, we can parallelize almost everything, but sometimes it makes no sense, like in my case. It seems it is not useful if you try to index some data in parallel.
I am going to continue my experiments, provide more sophisticated validation, make a simple http server and browser UI for my application, and save more information about the synthesis. I will try to make an automatic integration of the application into my working process.
I liked Golang, and it was nice to discover that almost any engineer can enter and make simple applications without a huge effort to learn the language. Golang is simple, succinct, but powerful.